All protein-protein interactions are defined by a binding equilibrium, an association (Ka) and dissociation (Kd) rate. This rate is determined by the 3D structure of the specific region on protein surface that interacts or binds to another biomolecule, known as the epitope. Mutations in the epitope amino acid sequence, or mutations elsewhere that alter the protein’s 3D structure, change this epitope and therefore the binding equilibrium. Changes in protein binding interactions alter function, and therefore ultimately determine the state of health or disease.
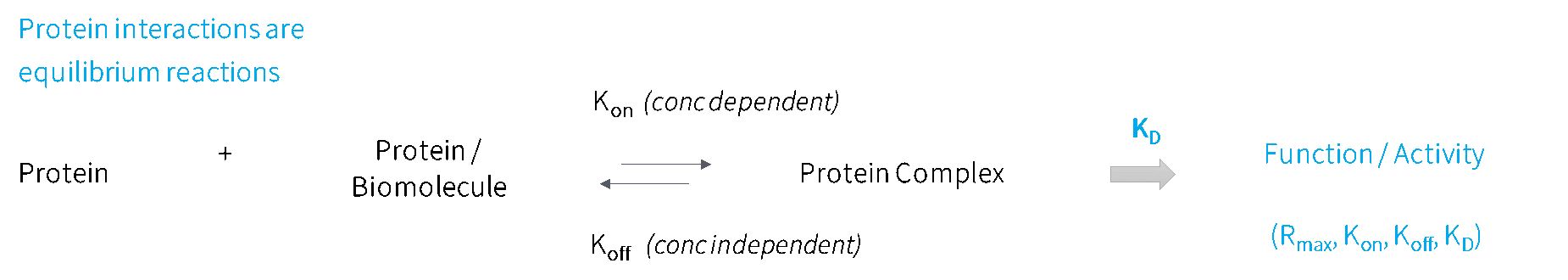
Changes in protein binding kinetics have been shown to be associated with antibody-dependent cellular cytotoxicity (ADCC) efficiency (Tan et al, 2022; Bruhns, 2012), cancer immunotherapeutic efficacy (He et al, 2017), pathogen mutational therapeutic drug and vaccine escape (Wang et al, 2022; Tuekprakhon et al, 2022; NIH COVID-19 Treatment Guidelines), autoimmune disease severity (Egner, 2000; Goilav & Putterman, 2015; Suwannalai et al, 2014; Rombouts et al, 2016), to name a few. This highlights the importance of measuring kinetics at scale. Something uniquely offered by SPOC biosensors.
Using real-time, label-free kinetic data collection with SPR sets SPOC apart from other proteomic arrays on the market which utilize end-point assays, such as fluorescence. We compared two commercial antibodies targeting the fusion capture tag using an end-point fluorescent assay and SPOC SPR assay and found that the anti-capture tag antibody with high-off rate (left) was missed by the fluorescent assay, while SPR detected binding events for both antibodies. This demonstrates how critical the method of data collection is to the outcome of your research.
SPOC can simultaneously collect qualitative, quantitative, and kinetic data for up to 1000 individual proteins at once. Acquisition and analysis software is in development to enable up to 2,400 individual proteins simultaneously. SPOC’s unique capability to measure differential kinetics combined with customized chips containing customer-defined proteins of interest facilitates a number of applications, including:
Contact us to ask about your desired application!
Egner, W. The use of laboratory tests in the diagnosis of SLE. Journal of Clinical Pathology vol. 53 424–432 Preprint at https://doi.org/10.1136/jcp.53.6.424 (2000) PMID: 10911799.
Goilav, B. & Putterman, C. The Role of Anti-DNA Antibodies in the Development of Lupus Nephritis: A Complementary, or Alternative, Viewpoint? Seminars in Nephrology vol. 35 439–443 Preprint at https://doi.org/10.1016/j.semnephrol.2015.08.005 (2015) PMID: 26573546.
Suwannalai, P. et al. Low-avidity anticitrullinated protein antibodies (ACPA) are associated with a higher rate of joint destruction in rheumatoid arthritis. Ann Rheum Dis 73, 270–276 (2014) PMID: 23463689.
Rombouts, Y. et al. Extensive glycosylation of ACPA-IgG variable domains modulates binding to citrullinated antigens in rheumatoid arthritis. Ann Rheum Dis 75, 578–585 (2016) PMID: 25587188.
He M, Chai Y, Qi J, Zhang CWH, Tong Z, Shi Y, Yan J, Tan S, Gao GF. Remarkably similar CTLA-4 binding properties of therapeutic ipilimumab and tremelimumab antibodies. Oncotarget. 2017 May 19;8(40):67129-67139. doi: 10.18632/oncotarget.18004. PMID: 28978021; PMCID: PMC5620161.
Tan R, Nie M, Long W. The role of B cells in cancer development. Front Oncol. 2022 Aug 11;12:958756. doi: 10.3389/fonc.2022.958756. PMID: 36033455; PMCID: PMC9403891.
Bruhns P. Properties of mouse and human IgG receptors and their contribution to disease models. Blood. 2012 Jun 14;119(24):5640-9. doi: 10.1182/blood-2012-01-380121. Epub 2012 Apr 25. PMID: 22535666.
Wang, Q., Guo, Y., Iketani, S. et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature 608, 603–608 (2022). https://doi.org/10.1038/s41586-022-05053-w
Tuekprakhon A, Nutalai R, Dijokaite-Guraliuc A, Zhou D, Ginn HM, Selvaraj M, Liu C, Mentzer AJ, Supasa P, Duyvesteyn HME, Das R, Skelly D, Ritter TG, Amini A, Bibi S, Adele S, Johnson SA, Constantinides B, Webster H, Temperton N, Klenerman P, Barnes E, Dunachie SJ, Crook D, Pollard AJ, Lambe T, Goulder P, Paterson NG, Williams MA, Hall DR; OPTIC Consortium; ISARIC4C Consortium; Fry EE, Huo J, Mongkolsapaya J, Ren J, Stuart DI, Screaton GR. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell. 2022 Jul 7;185(14):2422-2433.e13. doi: 10.1016/j.cell.2022.06.005. Epub 2022 Jun 9. PMID: 35772405; PMCID: PMC9181312.
NIH COVID-19 Treatment Guidelines: SARS-CoV-2 Variants Currently or Recently Circulating in the United States and Their Susceptibility to Anti-SARS-CoV-2 Monoclonal Antibodies. < https://www.covid19treatmentguidelines.nih.gov/tables/variants-and-susceptibility-to-mabs/> Accessed Jan 19, 2024.
1600 Adams Drive
Suite 236
Menlo Park, CA 94025
7201 E Henkel Way
Suite 285
Scottsdale, AZ 85255
© 2022, SPOC. All rights reserved.
Powered by: PixelbiteWT